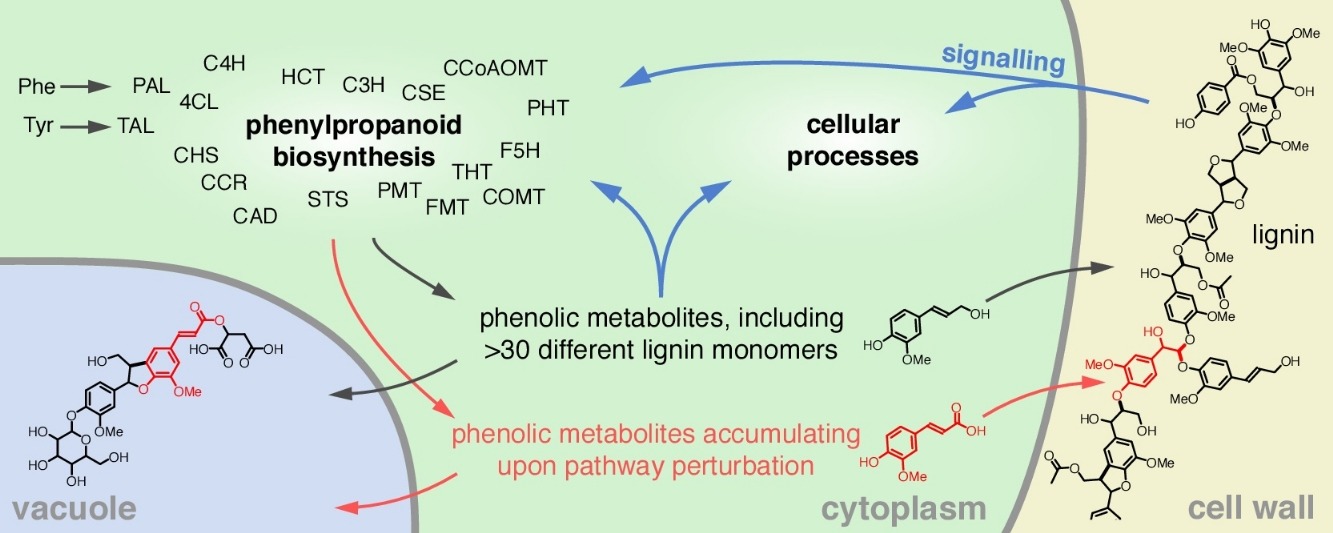
Identifying new genes in aromatic metabolism
When improving plant cell walls, it all comes down to identifying the genes that are involved in the biosynthesis of the major cell wall polymers, and altering their expression levels in target crops such as poplar and maize. Through co-expression analyses in Arabidopsis and maize, we have identified a set of candidate genes that likely play an important role in phenolic biosynthesis. We have already demonstrated the role for some of these in the biosynthesis of lignin and coumarins (Vanholme et al., 2013; Sundin et al., 2014; Vanholme et al., 2019). Our expertise in comparative metabolite profiling and mass spectrometry is a great asset to help elucidating their function. The potential for applications of these genes is investigated by analysing the biomass composition and saccharification potential of the corresponding mutants in Arabidopsis, poplar and maize. Higher order mutants are made to test for additive or synergistic effects (e.g. de Vries et al., 2018).